Genome editing
Team
Aleksandar Rakovic, PhD (group leader); Victor Krajka (PhD student); Britta Meier (research technologist)
Resources
The emerging CRISPR/Cas9 (clustered regularly interspaced short palindromic repeat) technology has been used to rapidly, easily and efficiently modify endogenous genes and genomic regions in a wide variety of different cell types and in organisms that have traditionally been challenging to manipulate genetically.
The system comprises of i) a short synthetic RNA (gRNA) composed of a “scaffold” sequence necessary for Cas9-binding and a user-defined ∼20 nucleotide “spacer” or “targeting” sequence which defines the genomic target to be modified and ii) the endonuclease Cas9. Upon recognition of the genomic target sequence by the gRNA, Cas9 cleaves each of the targeted DNA strands.
The resulting DNA breakages will be repaired by the error-prone non-homologous end joining (NHEJ) pathway resulting in random insertions or deletions (InDels) inducing the frame shift and finally knocking out the targeted gene. Alternatively, Cas9-mediated DNA cleavages can be repaired by the homologous recombination (HR). By providing a donor DNAs one can introduce desired mutations or correct the existing ones genome wide.
The CRISPR/Cas9 genome editing platform is fully established within the Dr. Rakovic’s research group “Molecular mechanisms of Parkinson’s disease” at the Institute of Neurogenetics. His group has already successfully established ~20 CRISPR/Cas9-derived models of Parkinson’s disease and Dystonia using induced pluripotent stem cells (iPSC) and commonly used cell lines.
The CRISPR-Cas9 system is a game changing technology for precise and efficient alterations in genome sequence and gene expression. The accessibility of this novel genome editing technology will fundamentally improve and influence biological/medical research in the elucidation of molecular mechanisms as well as the development novel molecular therapeutics for human disease.
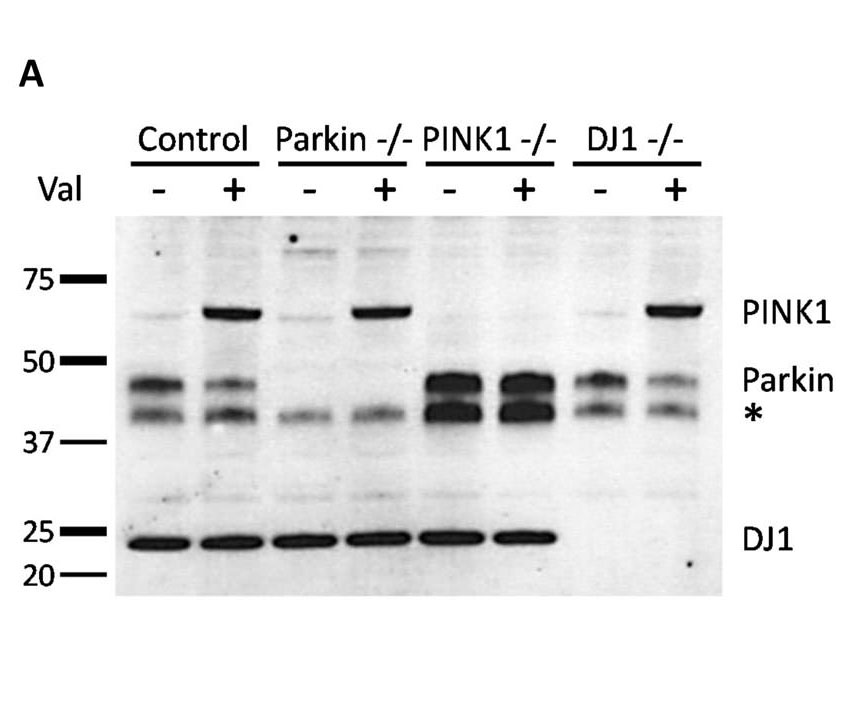
Generation of Parkin, PINK1, and DJ1 knockout isogenic neuroblastoma lines using CRISPR/Cas9. Western blot analysis using antibodies against Parkin, PINK1, and DJ1 shows complete knockout of targeted genes.
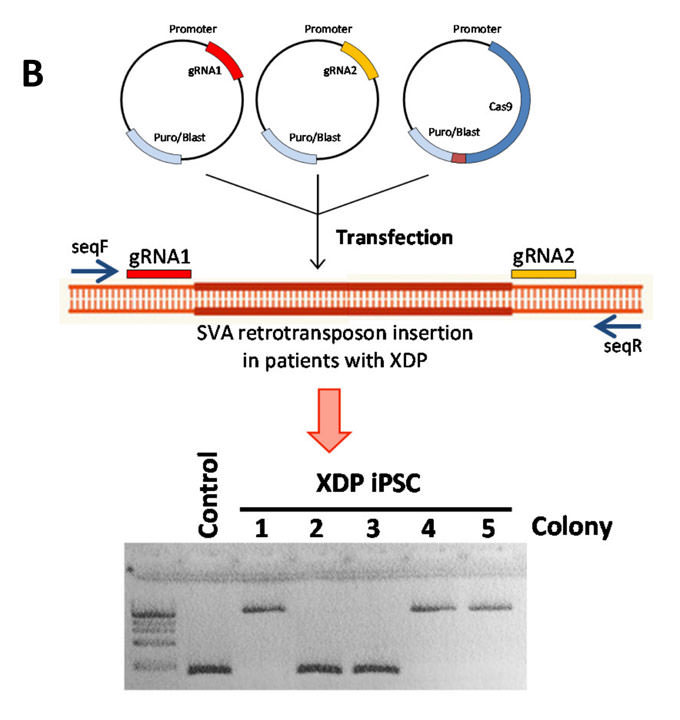
CRISPR/Cas9-mediated removal of a large intronic insertion (SVA) in intron 32 of the Taf1 gene in iPSC derived from the XDP patient. PCR using “outer” primers seqF and seqR shows correction in two (clones 2 and 3) out of five analyzed clones.
