Genome Data Analysis Core (GDAC)
A central aspect of ProtectMove is the generation of large-scale genomics data using a range of different high-throughput methods. The mere volume and complexity of the data generated by these powerful technologies is constantly increasing, creating a need for extensive bioinformatics expertise and a dedicated high-performance computing environment. To meet these needs within ProtectMove, we propose the creation of a dedicated Genome Data Analysis Core (GDAC) to serve as a centralized bioinformatics support facility for the primary research projects. GDAC will be embedded within the Lübeck Interdisciplinary Platform for Genome Analytics (LIGA) which was recently established within the Medical Faculty (‘Sektion Medizin’) at University of Lübeck and is affiliated with the Institute of Neurogenetics and the Institute of Integrative and Experimental Genomics. The overarching aim of LIGA is to excel genome research on campus with the goal of improving our understanding of the genetic and epigenetic foundations of genetically complex human diseases. All bioinformatics protocols and pipelines required for the analyses to be carried out by GDAC are either already in place or are currently being implemented by the LIGA team. This relates to many types of data (e.g. microarray-based, next-generation DNA and RNA sequencing) and involves many tasks (e.g. raw data processing, quality control (QC), mapping, variant calling, annotation, filtering and format conversion) throughout ProjectMove.
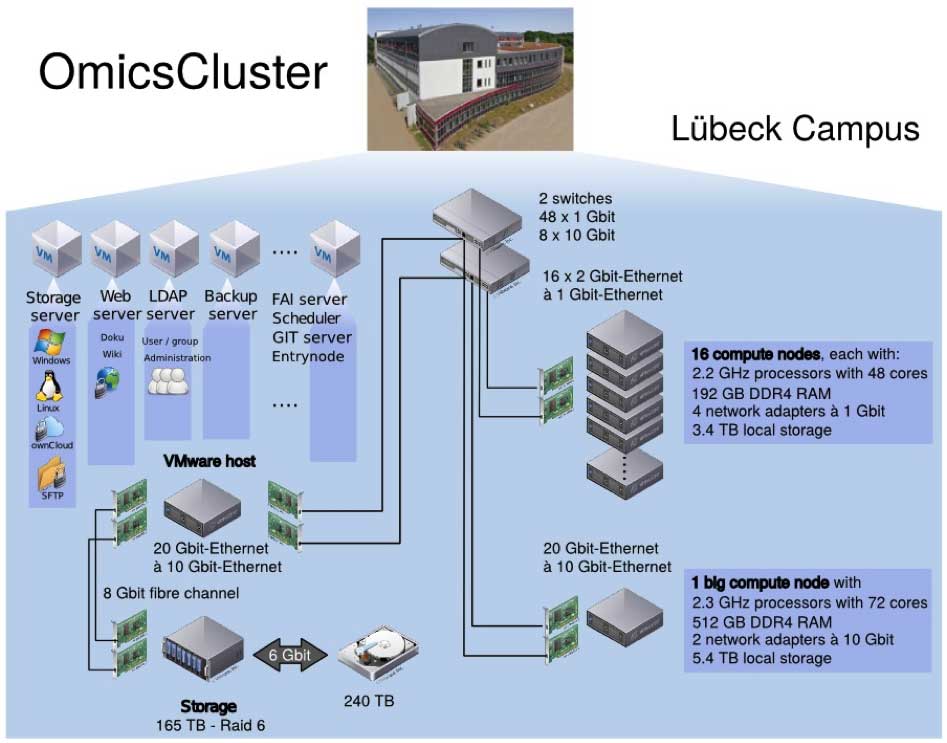
All GDAC operations and workflows will be implemented on a high-performance computing evironment (“OmicsCluster”) recently installed on the Lübeck campus (see Figure X). The cluster has been specifically tailored towards the analysis of high-throughput ‘-omics’ data and is currently being expanded further in terms of both its computational performance and storage capacity. In its currently installed configuration the OmicsCluster amounts to a compute capacity of 840 cores, 3.5 TB RAM and nearly 60 TB of local disc space. For long-term storage, an NFS storage system with 240 TB disc space is available that can be extended to several PB, if required, within the same architecture. Jobs on the system are scheduled using the Slurm software and data transfer is handled via ownCloud and command-line file transfer protocols (e.g. SFTP, Aspera). Version control of all programming code and pipelines is managed by a local Git server. The cluster is managed by a dedicated full time administrator who is also in charge of all hard- and software-related maintenance.
